Hi fellow steemians! Well, you probably noticed that great personal passion of mine is a technique called optogenetics. I had already written 10 articles on this topic and thus would like to put all the pieces together, so you can see the whole picture.
In short – optogenetics represent a fascinating method that combines optics and genetics with the aim to precisely control cells. Such approach was primarily used in neurobiology and basically you can control neurons and various brain regions of even living animals. So yes, mind control.

Source
In this article I will use information from my previous works and put them all together, but I will provide links to the original articles as well! The aim of the article is to give you an overview of how optogenetics work as well as to give a bit of a historical background to the method. This is a chance for those of you that did not read my original articles to get better understanding of the method. It is also going to be a bit longer, so fetch yourselves tee or something! :)
Happy reading!
Controlling mind with light
So, what is optogenetics you ask? Well, it is a relatively new method that combines optics and genetics to control the activity of cells, especially neurons, with spatio-temporal precision. This means that the level of control is acute both in space and time. And that represents a huge upside in comparison with electrical or chemical stimulation of neurons.
What is wrong with electrical stimulation? Well, even though electrical stimulation can be rapidly turned on and off (temporal precision) it has not defined borders of effect (as it travels in all directions). On the other hand, chemical stimulation of cells can be utilized in such way that it only affects specific cells (spatial precision), but the downside is that it can be irreversible or very slow.
For the aim of studying so dynamic and diverse structures such as neurons there is need for a fast, reversible and specific tool. Light is ideal in this case. It is very fast, you can turn it on and off and it will not zap other cells, but how can you do that?!
The basic steps
Key components of every optogenetic experiment are light sensitive proteins that change the activity of cells after light stimulation. The mostly used proteins are light-sensitive transmembrane ion channels called opsins. Opsins are subsequently expressed in neurons – that means that these neurons are genetically modified, so they produce these specific proteins in their cell membranes. This is usually done by inserting gene of interest (gene coding opsin) into viral vector and then infecting host cells with this virus (see the scheme bellow).
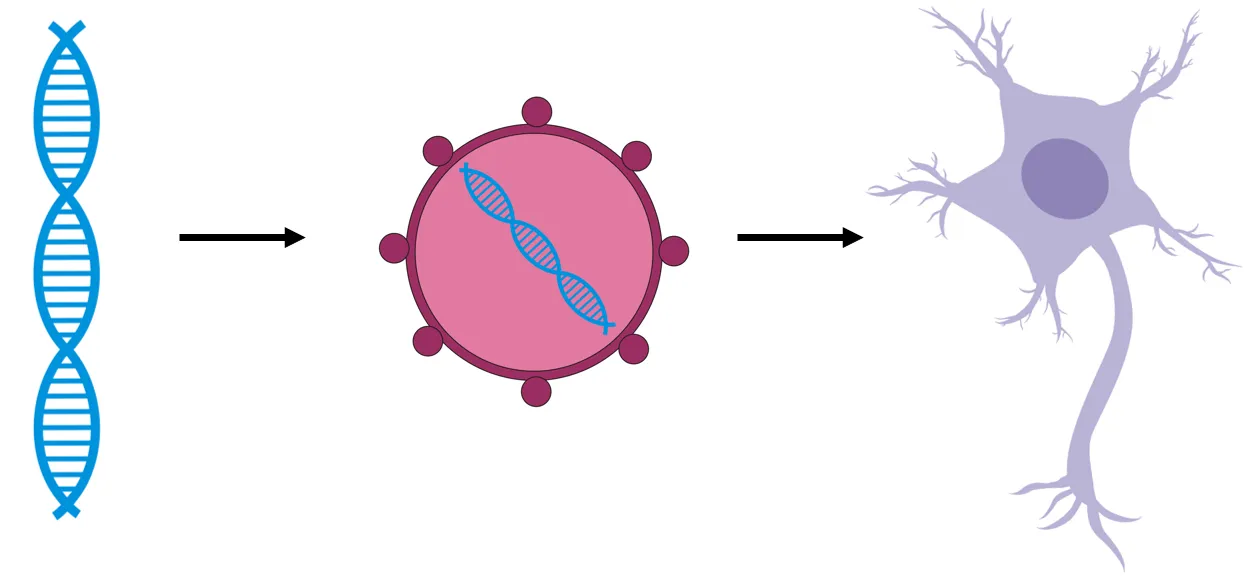
Gene of interest (the blue DNA) is inserted to neurons via viral vector.
Next step is light stimulation that causes conformational change of the opsin, which enables transportation of ions throughout the membrane and thus change in membrane potential. What does that mean? Well, change in membrane potential can be translated as change in concentration of ions inside and outside of neurons. Ions are charged particles meaning that when they move from one side of the membrane to another they change the charge of the membrane (either to positive or negative values). The change of membrane potential may result in either depolarization or hyperpolarization of cell membrane. Depolarization is synonym to activation of neurons whereas hyperpolarization corresponds to inactivation.
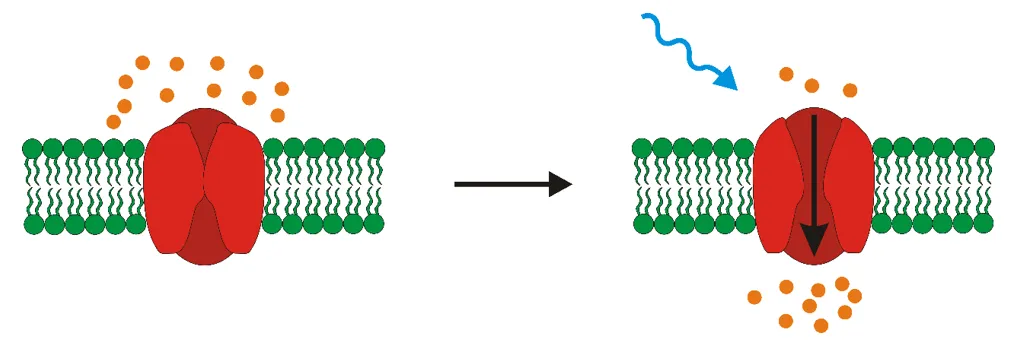
Light stimulation results in opening of the light-sensitive ion channel and subsequent change in membrane potential.
The whole process of achieving this is however a tad more difficult. But before we jump into that a little bit of historical overview and data.
History of optogenetics
Sometimes we find the answers we are looking for in unlikely places. Who would ever imagine that we will control brains with light?! Well, Francis Crick (Nobel prize winner and co-discoverer of double helical structure of DNA) did. In 1979 he proposed the idea to use light in neurobiology.
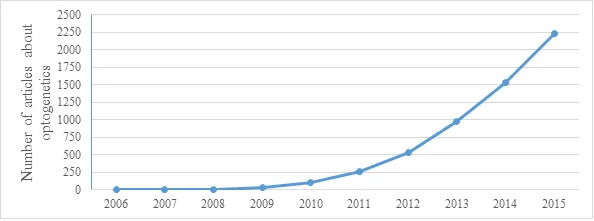
Unfortunately, the great gap between the study of the vertebrae brain and microbial proteins that the discovery of method such as optogenetics slowed down. Change came in 2005 - a year that marks the beginning of modern optogenetics. One of the light-sensitive proteins – Channelrhodopsin-2 (ChR2) from Chlamydomonas reinhardtii was expressed in neurons for the first time. Stimulation with blue light caused depolarization of these neurons. This technique was non-invasive and could be controlled in space and time. This was the corner stone for optogenetics (the method was titled optogenetics in 2006). Since then there was an explosion of scientific articles about optogenetics. Just look at the graph bellow that shows the increase in number of article in last years.
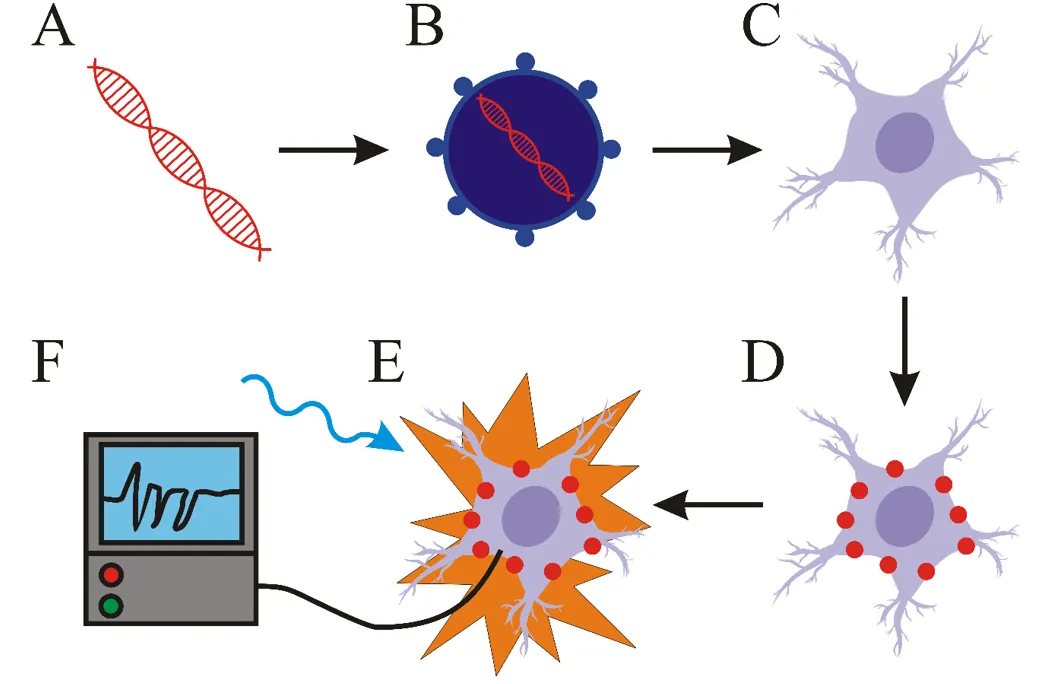
Key components of every optogenetic system
Back to the science part. The most important parts of optogenetic systems include light-responsive proteins (like opsins), a vector (viral vector for example) and a light source. As you read look at the scheme bellow. First step is the creation of a molecular construct carrying a gene for light-responsive protein (A). This construct is subsequently inserted into a vector (B). Cells transformed with this vector (C) express light-responsive proteins (D). The next step is light stimulation of transformed cells with light (E). The wavelength of the light has to correlate with the excitation spectrum of the light-responsive protein. The last step is the detection of the changes in activity of cell (F) (such as the change of membrane potential for example). For this purpose, various electrophysiological methods are most commonly used. Scheme bellow summarizes this, but we will take a closer look at each step!
Light-sensitive proteins
Most widely used light-responsive proteins are called opsins. For opsins to function properly, the presence of retinal is needed. Retinal, a form of vitamin A, forms a functional complex with opsin. Light stimulation of this complex results in photoisomerization of retinal which causes change in conformation of opsin.
Mostly used opsins are so called Type 1 opsins that originate from prokaryotes or algae. Type 2 opsins originate from mammals but have slower kinetics that are not favourable to artificially induce neuronal signalling.
Important thing now - how does actually neuronal signalling work? A neuronal signal is an electric impulse that is generated by the change of membrane potential on the inner and outer side of cell. The change of this membrane potential is caused by the change of concentrations of ions in intracellular and extracellular space via transport through ion channels. Well, and type 1 opsins, function exactly like that! They are bi-functional transmembrane proteins - not only capable or sensing and reacting to light but are also ion channels. Thus, light stimulation of these proteins results in their conformational change and flow of ions in or out of the cell - thus mimicking the exact same thing that neurons do!
Neurons can be either activated or inactivated. Activation is called depolarization and membrane potential has positive values in this case. Hyperpolarization equals to inactivation. Some opsins, such as channelrhodopsin-2 (ChR2), discovered in Chlamydomonas reinhardtii are capable of causing depolarization after light stimulation. The other widely used excitatory opsin is channelrhodopsin from algae Volvox carteri (VChR1). Others such as Halorhodopsin form Natromonas pharaonis (NpHR) could be efficiently used in cells to cause hyperpolarization. The mechanism of function and excitatory spectrum of these opsins is depicted bellow.
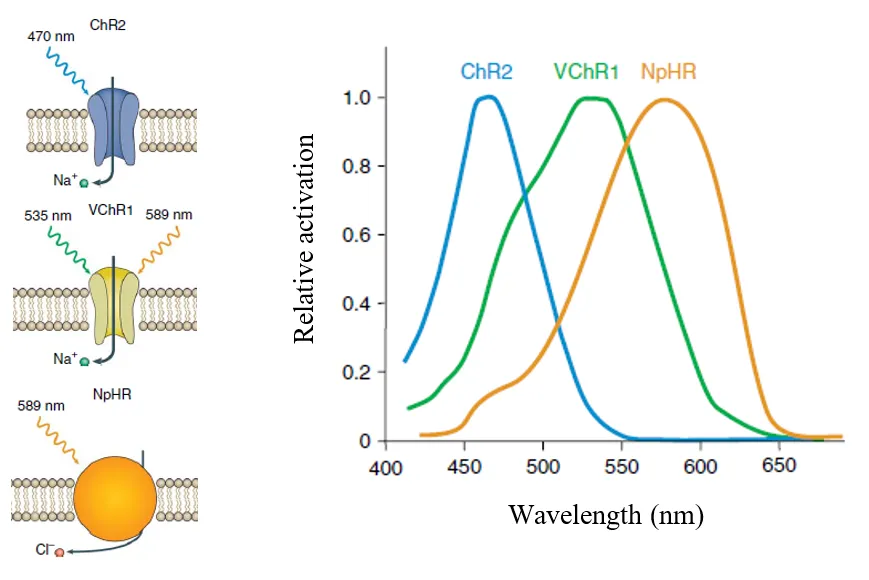
Mechanism of function and excitatory spectrum of ChR2, VChR1 and NpHr
Source
Gene delivery and expression
Expression
So, you have your microbial opsin that you want to put into the neurons. How to do that? Production of certain substances, mainly proteins, in cells is termed as gene expression. This means that the information coded by DNA is read by molecular machinery of the cells and transformed into product. Thus, we need to put a foreign DNA containing “recipe” for our opsin into the cell. More importantly we want to maintain the already mentioned spatial specificity of optogenetics – meaning that opsins will be expressed only in certain cells. Just a side-note – I also use the term transcription, that means that DNA is transcribed into RNA and that is subsequently translated into proteins – but basically you can just think about it as an equivalent of gene expression.
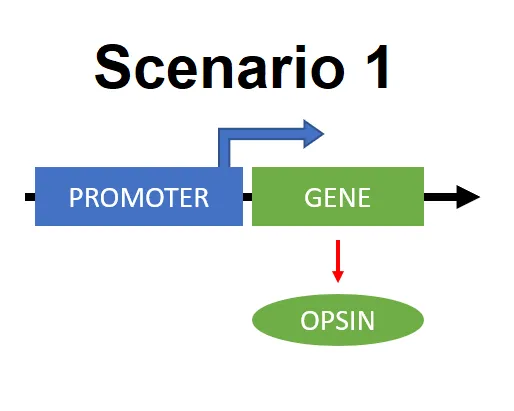
This specificity is due to the usage of cell specific promoters - simply regions of DNA that initiate expression of certain gene. There are more ways to achieve this. One way is that the expression of opsin can be directly under the control of promoter that is active only in certain cells. (Scenario 1)
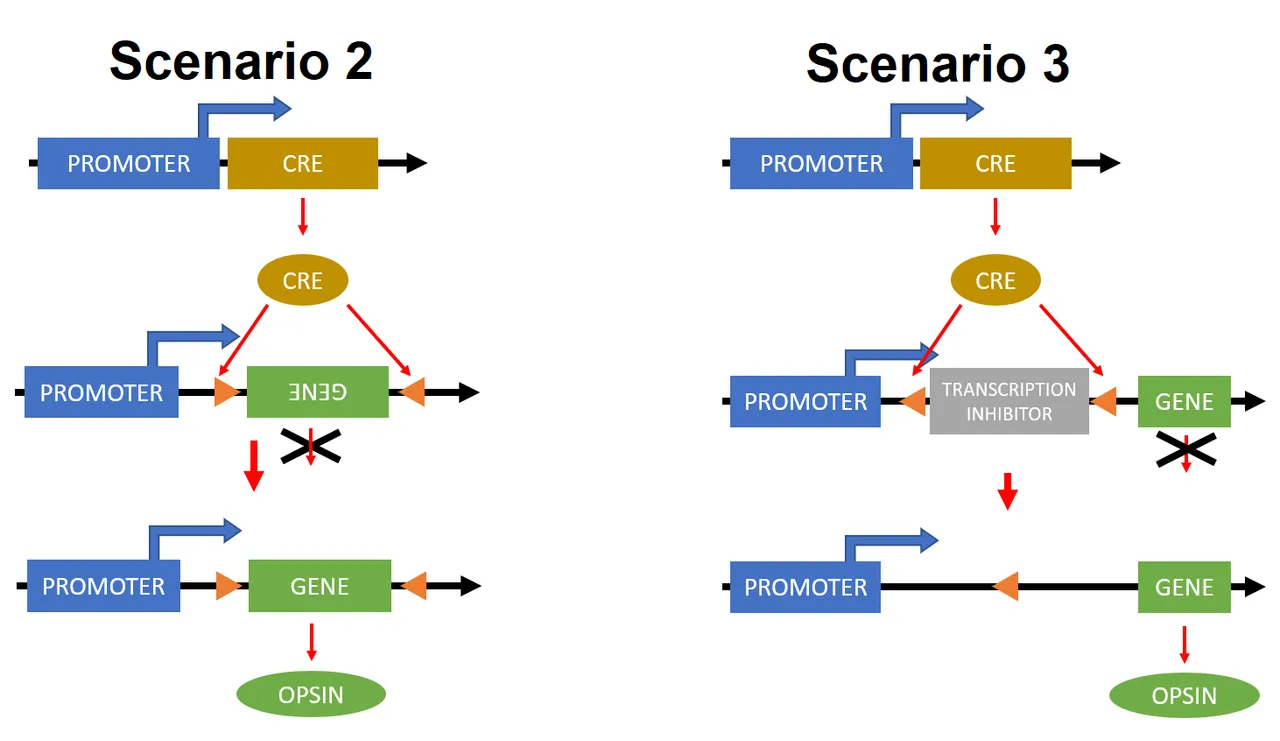
Another approach is the use of binary systems. These are composed from two parts – driver and reporter genes. The expression of driver genes is directly under the control of cell specific promoters. Reporter genes are active only in cells in which are driver genes expressed. An example of a binary system is the usage of Cre recombinase. It catalyses the recombination between two loxP sites (specific homological sequences of DNA). Just to clarify, recombination means that two regions of DNA exchange their parts recombine and then they are mixture of one-another. Recombination causes either deletion or inversion of DNA localized between these two loxP sites (it all depends on the orientation of loxP sites). The gene coding Cre recombinase is expressed only in specific cells – Cre is termed driver gene in this case. Now we have two possible scenarios. In one of them our reporter gene, an inverted opsin-coding gene, is bounded between two lox sites (orange triangles). When Cre recombinase is presented, then this gene gets into correct position and opsin is being expressed (Scenario 2). Another way of using binary systems is to put an inhibitor of transcription between loxP sites - see they are aligned a bit differently. When Cre recombinase is expressed in such cells the inhibitor is cut out of the DNA region and the transcription may happen and thus opsin is being expressed (Scenario 3 in the scheme bellow).
Delivery
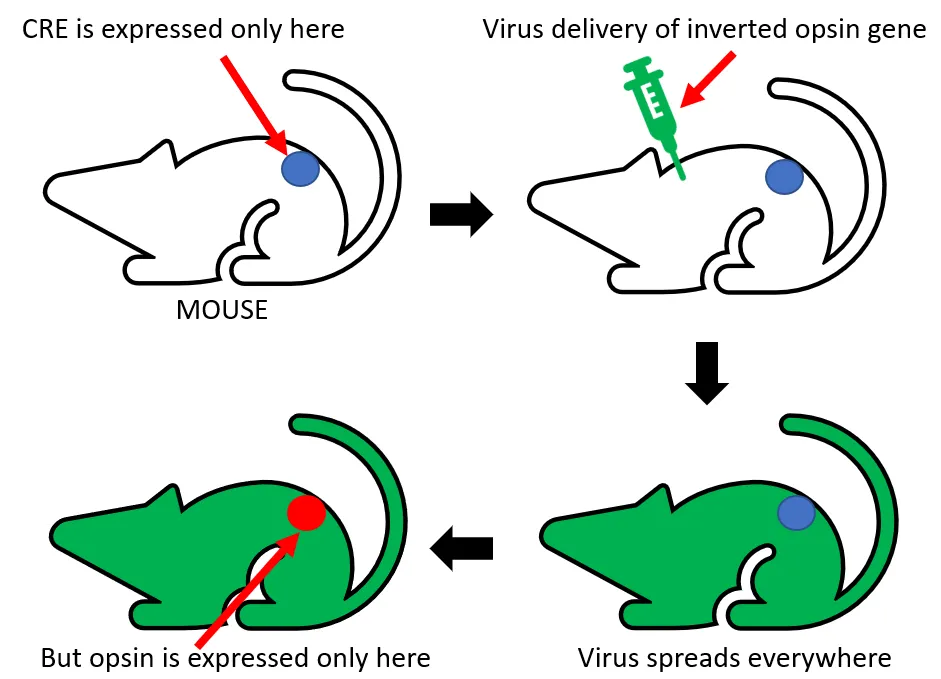
There are generally three ways of transportation of genetic constructs to cells of interest. The first is the usage of viral vectors. The expression of proteins is fast and robust. The downside of this strategy is the limited amount of DNA that can be put into a virus. Transgenic animals represent a second option of vectors. Using this approach allows the insertion of larger DNA sequences. The main limitations include the time-consumption of this method and limitation of usable organisms. The third and probably the most effective strategy combines the previous two. It is tightly bound to binary systems. Driver gene coding Cre recombinase has to be expressed only in specific locations of transgenic organism. The virus will carry the reporter gene - the inverted opsin sequence, bounded by two loxP sites. Even though the virus will infect all cells, opsins will be expressed only in cells with driver genes. This strategy combines the high spatial specificity of transgenic approach and the option to use several different opsins in one organism, thus bypassing the time-consuming process of creating a new transgenic line. This might also be kinda hard to understand so again I will try to show you this last strategy on schematics bellow.
Light delivery
Now that the we have cells expressing our light sensitive proteins we need to stimulate them with light. How to perform such feat? For experiments in vitro where you work only with cells there is no problem with light delivery - you can just shine light onto the cells. For experiments in vivo on living animals it is a bit difficult. The most common strategy is the integration of optic fibre with the brain of the rodent (remember - we are stimulating neurons and thus brain). Similarly, as electric wires transport electricity from one place to another, so does optical fibre transport light. This fibre is inserted into the skull of lab animals, mainly rodents, so specific parts could be illuminated. New alternative to optical fibres are wireless devices that use LEDs – but still, they are quite bulky. This brings us to the question of source of light. There are two options- lasers and LEDs. The choice is highly dependent on available finances and the method of usage. Lasers work well when coupled with fibre optics for stimulation in vivo (meaning in living animals), but they are expensive and fragile. On the other hand, LEDs are cheaper and more durable, but they are inferior in power and also emit excessive heat that may flaw results of experimentation.
When talking about using light in optogenetics I would like to mention some key properties of light stimulation that have to be upheld for successful application of optogenetics. Firstly, the wavelength of used light has to correlate with the excitatory spectrum of opsin. This means that the opsin will change its conformation only when correct wavelength is applied and thus, only then changes the membrane potential of cell. And secondly, the right frequency of light pulses has to be applied. This means that in order to achieve successful production of artificial neuronal signals you need to activate opsins with correct frequency!
Interpretation of results
Yay! You have successfully done your optogenetic system and everything works – right? Hell, how do you have to know without proper methods of interpretation of the results of your optogenetic experiments. There is necessity of existence of methods capable of localization of opsins in cells or organisms, but also methods that enable to monitor the change of membrane potential after light stimulation. Interpretation of results from these experiments enables us to better describe the properties of opsins, but also helps to optimize experimental conditions.
Expression of opsins in cells is mostly visualised via the use of fluorescent proteins. Genes coding these proteins are part of the constructs that contain genes for opsins and so this fusion enables the localization of cells for further experiments. Commonly used fluorescent proteins include GFP, YFP or mCherry. Basically you use fluorescent microscope and cells that glow are those that express your gene of interests. Next step might be to test the properties of cells and opsins. For this a method termed patch clamp is most commonly used. Patch clamp is a electrophysiological technique that enables to measure changes of electric current or voltage which are created as result of ionic flow through membrane and thus change of membrane potential. Essential component of this method is a glass micropipette, whose tip has a diameter of only several micrometres. Electrode residing inside the pipette is capable to measure even small changes of electric current. In order to correctly measure these changes, the formation of a so called giga seal is necessary. This is achieved by applying positive pressure to form a tight connection between cell membrane and tip of the pipette- so there is no leak current. There are different variations of patch clamp technique and type of the experiments determines which one should be used. Previously mentioned principle is used in cell-attached patch (A)- this enables to measure activity of one or more ion channels without destroying the integrity of cell membrane. Another variation, the whole-cell patch (B), enables the detection of activity of all channels in cell. The only difference from cell-attached patch is the application of strong suction after formation of giga seal- that causes tearing of small part of cell membrane and so the inside of the cell is directly connected with the solution and electrode inside the pipette.
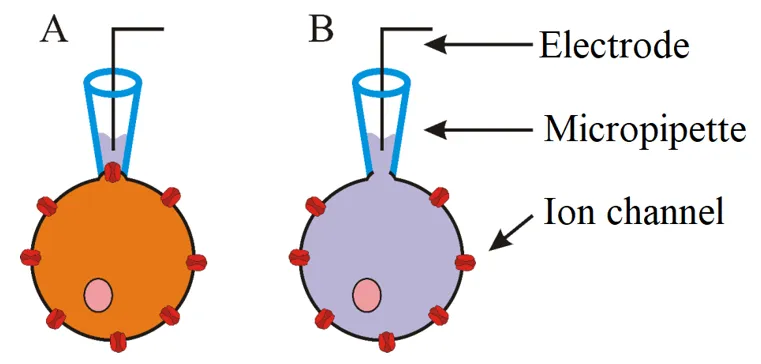
A - Cell-attached patch, B - Whole-cell patch
Such approach is however used only in in vitro experiments. With living animals, you could use behavioural testing. For example, optogenetic activation of certain brain regions in mouse activated or inhibited aggressive behaviour or motor activity. This method of evaluation is not very quantitative in nature and it is hard to visualize what really happens in the brain. On the other hand, many times the results are quite obvious. See demonstration bellow.
Conclussion
Optogenetics represent a versatile and modifiable approach for controlling and dissecting neuronal pathways with spatial-temporal precision in normal or pathological tissue. So far it has been applied to control activity of neurons in vitro and also in vivo to understand how activity of certain neurons affects behaviour. Even though an optogenetic approach is well applicable for understanding and perturbing neuronal functioning, there is still much to be done. This includes the development of new light-responsive proteins or the optimization of functioning of already developed ones. There is need to create new vectors, more efficient and compact light sources, or effective methods for the interpretation of results. Optogenetics, primarily a neurobiological tool, has already been used to control the activity of heart cells or intracellular signalling. This method opens a door to completely new approach of biological study. The usage of light and the potential it encompasses as non-invasive, fast and precise tool promises progress in its use. The future is always a mystery, but the upcoming years should show exciting progress in the development and application of optogenetics to deconstruct the functioning of cells and help us answer prominent question that lingers here as long as humanity: How?
Thank you for your time!
Previous articles about optogenetics:
Part 1 – Introduction
Part 2 – History of Optogenetics
Part 3 – Opsins – light sensitive proteins
Part 4 – Gene expression and delivery in Optogenetics
Part 5 – Light stimulation and delivery in Optogenetics
Part 6 – Readout of results in Optogenetics
Part 7 – My own research experience of using Optogenetics
Part 8 – Activate genes with light
Part 9 – Fighting cancer with light
Part 10 – Controlling hearts with optogenetics
References:
Mainly research and reviews from Karl Deisseroth, M.D., Ph.D. - leading expert on optogenetics
Other references directly in the text (some more than once, sou you do not have to scroll for the term! :) )
I also took information from my previous articles (list above) so for further information go and check those.
P.S.
Would you like more articles about optogenetics? :) Tell me in the comments!