
Here it is, the long-awaited post on CRISPR/Cas9! Please read my introduction post about nucleases first, as I won’t explain the things covered there again.
In the other post, I already introduced you to nucleases, DNA double-strand breaks and repair mechanisms for these breaks. These essential elements are the same, the main difference between TALENs/ZFNs and the CRISPR/Cas9 system is how the region where the break is introduced is found, and how the DNA is cut.1
But let’s take this from the start.


Just like the other gene-editing methods, CRISPR/Cas9 didn’t just fall out of the sky. It already existed in nature and was discovered and then modified by scientists. “CRISPR” stands for clustered regularly interspaced short palindromic repeats, while “Cas” means CRISPR associated. In 1987, these repeats were discovered in Escherichia coli and Salmonella typhimurium, both bacteria.2 It was later found out, that CRISPRs are found in many bacteria and archaea3 and serve as a defense against phages4, which are viruses that attack prokaryotes (don’t have a nucleus) but not eukaryotes (have a nucleus). @suesa
This defense looks a bit like this:
Viral DNA enters the cell and is recognized by a Cas protein thanks to a so-called protospacer adjacent motif (PAM) with a length of 2 - 6 nucleotides5. The sequence next to the PAM, the protospacer then serves as a template for a spacer that’s integrated into the cell’s own genome between regular repeats.6
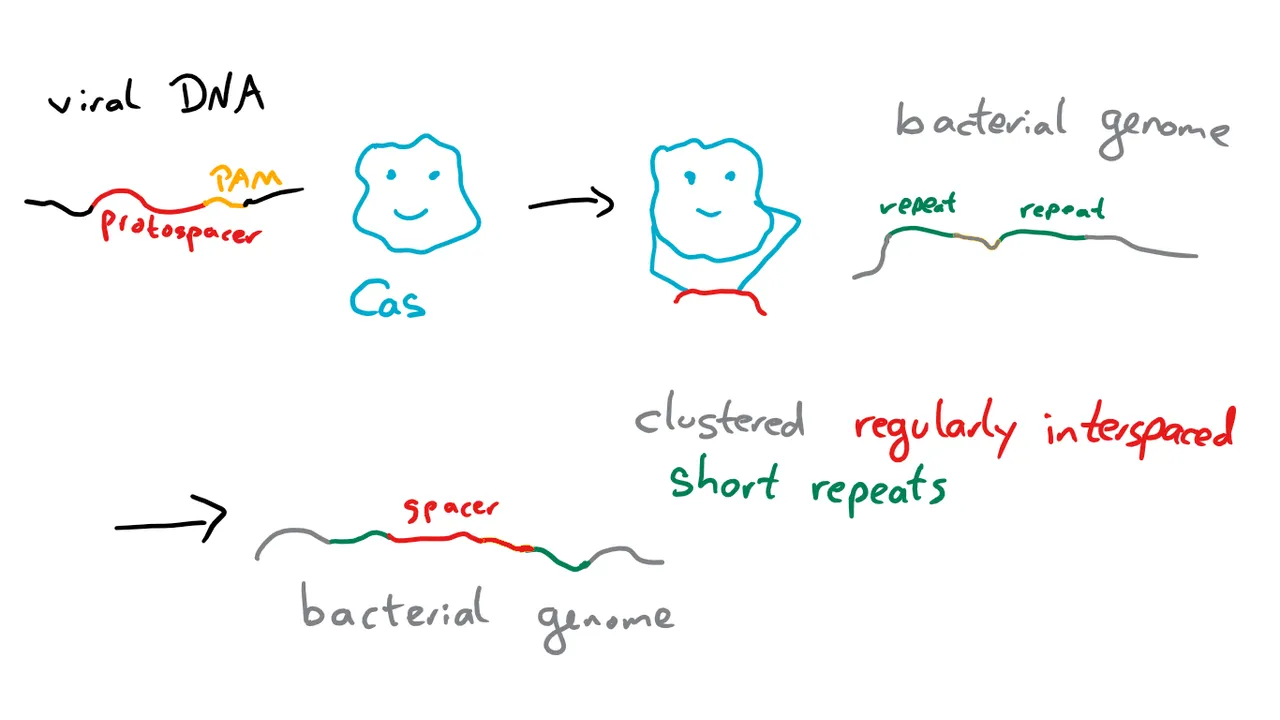
The next time the DNA of a phage like this enters the cell, it’s recognized by another Cas protein thanks to a combination of trans-encoded small CRISPR-RNA (tracrRNA), that is specific to the Cas protein, and a guide RNA (gRNA), which is specific for the DNA sequence targeted. The Cas protein then cuts the DNA and renders it harmless. Or at least that’s one of three slightly different way CRISPR/Cas can work.7. I’m not going to explain the other ones in detail, as this is the one relevant for CRISPR/Cas9. But if you want, you can read source 7.
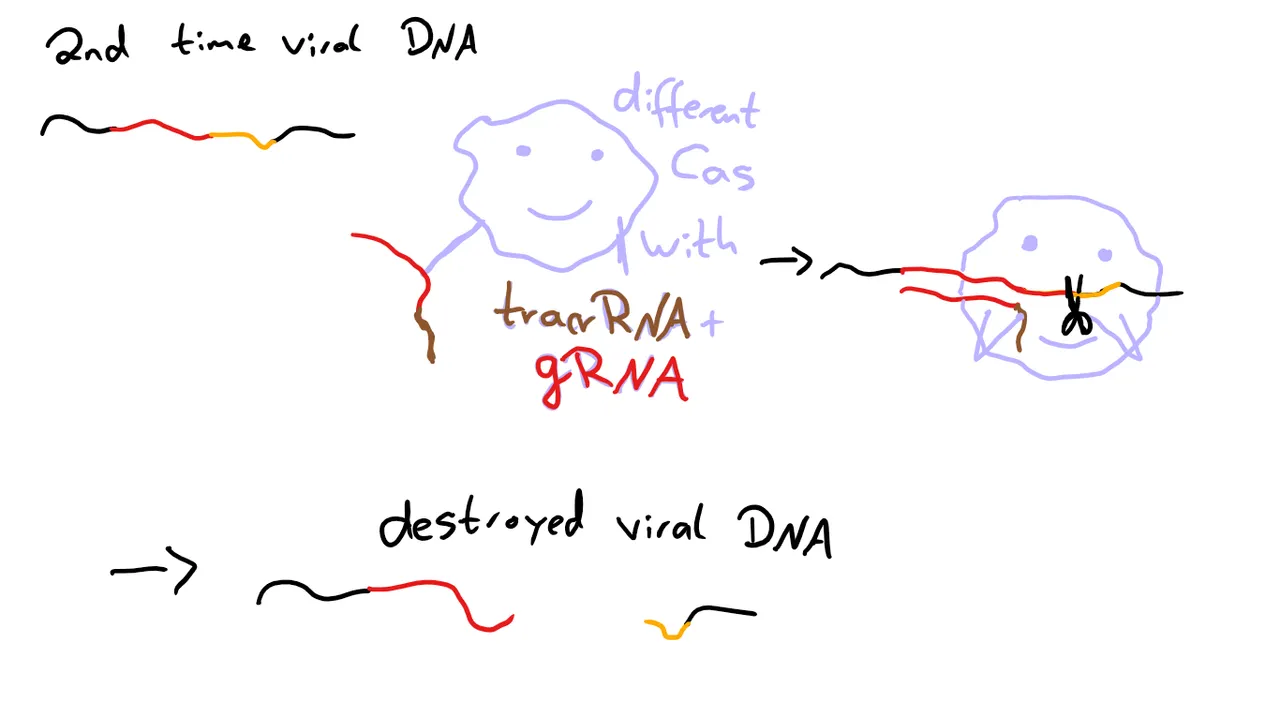


Okay, so, we have this exciting immune defense in bacteria, but how do we use it to edit genes?
It’s actually comparably easy.
The tracrRNA and gRNA I mentioned before can be synthesized in the lab, as both are very short. The gRNA can be adjusted to match any desired sequence, as long as it is next to a PAM8. The PAM sequence of Cas9, which is the endonuclease that belongs to Streptococcus pyogenes, reads “NGG” - random nucleobase, Guanine, Guanine.9 You can probably imagine that, with only 4 nucleotides available (Adenine, Guanine, Cytosine, Thymine), this sequence isn’t that rare.
Now, compare it to the ZFNs and TALENs from the last post: They need a lot more to find a specific DNA sequence. And that’s what makes CRISPR/Cas9 such a great tool; it’s a lot easier to design a gRNA than to develop a ZFN, which matches the target sequence in several base triplets.
But
Having a PAM that frequently appears in the genome doesn’t come without its danger. In the unfortunate case that the sequence appears more than once in the genome, together with a PAM, Cas9 might cut in the wrong place, which generates an off-target hit (= a cut in a location where it is not supposed to be).
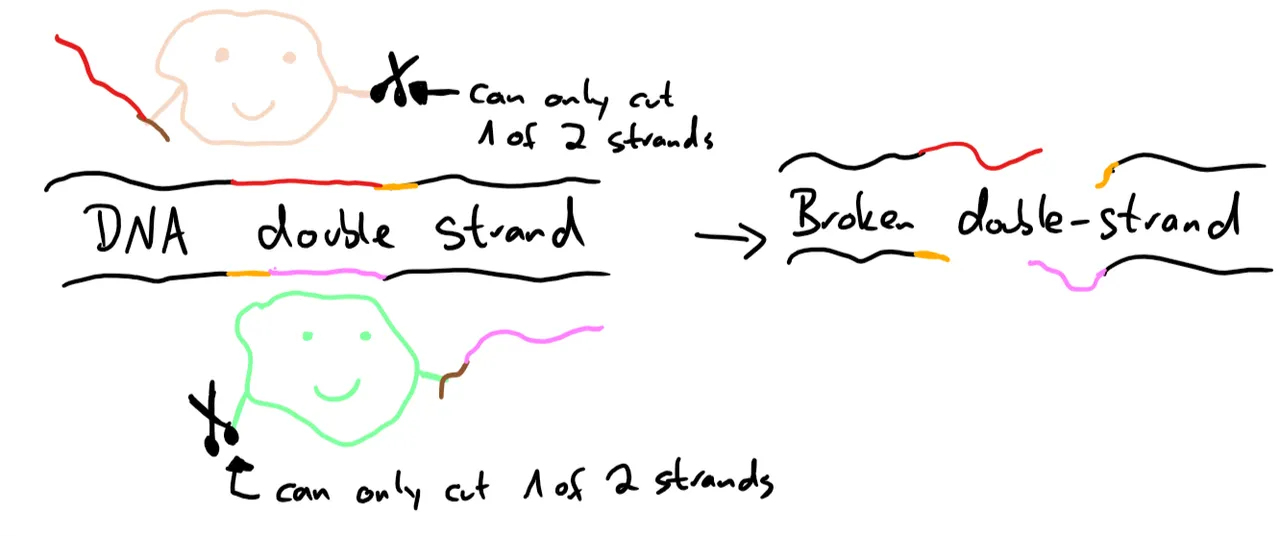
To circumvent this, Cas9 can be converted into a nickase. Usually, Cas9 has two parts that each cut one of the DNA strands, to introduce a double-strand break. But if you deactivate one of those, there is no complete break. Now introduce two Cas9 nickases which each have a different gRNA which causes them to cut in similar locations, but in different strands, the break can only happen if both nickases cut in the correct position. A wrong cut is immediately fixed by the cell’s repair mechanisms.10

There is, of course, a lot more to know about CRISPR/Cas9. There are ebooks on how it works exactly and how to use it that have over 100 pages (you can download one here for free if you want. Yes, I’ve read that), there are publications over publications and experiments that combine CRISPR/Cas9 with other methods, trying to improve it further.
Because despite how great it sounds, it’s still a lot of work and does not work in 100% of cases, by far not. Like every biological process, in the end, it’s statistics. Enzymes don’t know what they’re doing, they can’t think. It’s impressive that anything in nature works as it is supposed to if I’m really honest.
CRISPR/Cas9 is a new technique, we only really know how to use it since 2012, and that is not a long time, not in biology. Still, we’ve made progress, huge progress! And I am really looking forward to what the future brings, as there are so many possibilities we might not even have thought of yet.
Sources:
1 Cong, L., et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science, 339(6121), 819-823. doi: 10.1126/science
2Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., & Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology, 169(12), 5429-5433. doi:10.1128/jb.169.12.5429-5433.1987
3Jansen, R., van Embden, J. D., Gaastra, W., & Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology, 43(6), 1367-1681. doi:10.1046/j.1365-2958.2002.02839.x
4Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., & Soria, E. (2005). Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. Journal of Molecular Evolution, 60(2), 174-182. doi:10.1007/s00239-004-0046-3
5Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., & Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology, 155(3), 733-740. doi:10.1099/mic.0.023960-0
6Bhaya, D., Davison, M., & Barrangou, R. (2011). CRISPR-Cas Systems in Bacteria and Archaea: Versatile Small RNAs for Adaptive Defense and Regulation. Annual Review of Genetics, 45, 273-297. doi:10.1146/annurev-genet-110410-132430
7 Makarova, K. S., et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nature Reviews Microbiology, 9(6), 467-477. doi:10.1038/nrmicro2577
8Jinek, M., Chylinksi, K., Fonfara, I., Hauer, M., & Doudna, J. A. (2012). A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science, 337(6096), 816-821. doi:10.1126/science.1225829
9Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangoi, R., Horvath, P., & Siksnys, V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research, 39(21), 9275-9282. doi:10.1093/nar/gkr606
10Shen, B., et al., (2014). Efficient genome modification by CIRPSR-Cas9 nickase with minimal off-target effects. Nature Methods, 11(4), 399-402. doi:10.1038/nmeth.2857
